Neutron crystallography is an important complementary technique to X-ray crystallography since it provides details of the hydrogen atom and proton positions in biological molecules. Furthermore, as neutrons are a non-destructive probe, the resulting structures are free from radiation damage even at room temperature.
[adsense:336x280:8701650588]
Knowledge of H-bonding networks, water molecule orientations and protonation states, along with details of hydrophobic and electrostatic interactions, can prove vital towards a better understanding of many biological processes, such as enzyme mechanisms and can help guide structure-based drug design.
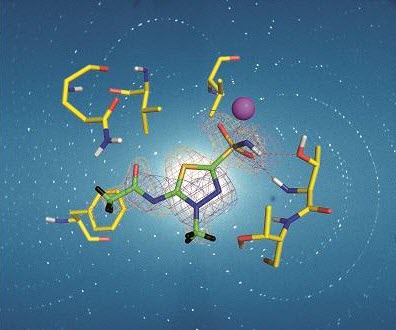
The first neutron crystallography study of a clinically used drug bound to its target was that of acetazolamide (AZM), a sulphonamide, which binds with high affinity to human carbonic anhydrase isoform II. Human carbonic anhydrases (hCA) are zinc metalloenzymes that catalyse the interconversion of CO2 and H2O to HCO3- and H+, an important reaction for many physiological processes including respiration, fluid secretion and pH regulation. As such hCA isoforms are prominent clinical targets for treating various diseases, such as glaucoma and epilepsy. hCA II is one of 12 catalytically active isoforms and, due to sequence conservation between them, substantial off-target binding to other isoforms occurs, reducing drug efficiency and causing side effects. Hence, there is a need to design effective hCA isoform-specific drugs. Over 400 X-ray crystal structures have been determined for hCA II, with around half of these in complex with inhibitors, yet despite the large amount of X-ray structural data available, key details regarding the H-atom positions of the protein and solvent and the charged state of the bound inhibitor were, until recently, missing for all but the hCA II/acetazolamide complex.
In the September issue of IUCrJ [Aggarwal et al. (2016), IUCrJ. 3, 319-325; doi:10.1107/S2052252516010514] McKenna and co-workers describe X-ray and neutron crystallographic studies of hCA II in complex with the inhibitor methoazolamide (MZM) providing missing details of the H-bonding and hydrophobic interactions in the complex, and identifying the charged state of MZM. They then compare the binding of AZM and MZM in the room-temperature neutron structures and discuss the observed differences in binding in terms of the enthalpic and entropic contributions to drug binding, suggesting that in the case of MZM, hydrophobic forces perhaps compensate for the loss of an extensive H-bonding network.
Over the past few years, a growing number of neutron structures have been deposited in the Protein Data Bank, including a number of other examples of enzyme-drug complexes. Although the overall number of neutron structures is still relatively small, there are growing numbers of examples for which neutron crystallography has provided the answers to questions that have remained elusive using other techniques.













