About Authors:
Sanjay Kumar Yadav*1, Anjana Yadav1, Shahana Majumder2
*1Dept of Pharmaceutical Chemistry, B.B.S. Institute of Pharmaceutical & Allied Sciences, Greater Noida, (U.P.), India
1Dept of Biotechnology, Chhatrapati Sahu Ji Maharaj University, Kanpur, (U.P.), India
2Dept of Biotechnology, Sharda University, School of Engineering and Technology,
Greater Noida, (U.P.), India
*sanjay_yadav3333@yahoo.co.in
ABSTRACT
Viruses are pathogens with an extremely narrow host range. Their phylogenetic origin is vague, tough it has always been considered that viruses are genes that became vagrant after having excluded themselves of the host’s or a related species’ genome (Anderson et.al., 2004). Viruses are usually units consisting of nucleic acids and coat proteins called capsids. Viroids consist only of RNA, i.e. they contain no protein at all. Except for a few cases, Viruses have no energy metabolism of their own. Consequently, they cannot perform syntheses and are thus unable to replicate themselves. Depending on their host species, it is distinguished between plant viruses multiplying almost exclusively within plant cells, bacterial viruses (bacteriophages) that depend on living bacteria, and animal viruses. The genetic information of viruses is either encoded by single-stranded RNA (Most plant viruses), double-stranded RNA (Wound tumor viruses), single-stranded DNA (Gemini viruses) or double stranded DNA (Cauliflower mosaic virus). Based on the shape of the viruses particle, it is distinguished between rod shaped and icosahedral viruses with a capsid that seem almost spherical. Viruses’ cause many disease in humans of international importance for example smallpox polio, hepatitis etc. viruses cause also many important plant disease and are responsible for huge loss in crop production and quality in all part of the world. Around 25 years ago, when genomic properties of geminiviruses were studied, many scientists regarded them as ‘friends of humans’, because of their potential as gene transfer vectors in plant genetic engineering and non-harmful effects on host plants. But far from being friends, these viruses have now emerged as foes and are a serious threat to world agriculture now. Increase in international commodity trade, intercontinental transportation networks and a changing global climate have contribute to the spread of this virus and its whitefly vector (Moffat et.al.,1999).
[adsense:336x280:8701650588]
Reference Id: PHARMATUTOR-ART-1377
INTRODUCTION
Plant viruses are divided into more than 15 families, of which Geminiviridae constitutes the second largest family after the Potyviridae besides their importance as plant pathogens. Gemini viruses are ideal models for the study of replication and gene expression in plants, due to the fact that they replicate in the cell nucleus via a dsDNA molecule1. Such studies have generated a large amount of information regarding the molecular biology of geminiviruses. Gemini viruses are characterized by their twin geminate particles, ssDNA circular genomes and whitefly (Bemisia tabaci) mediated transmissibility. Geminiviridae is represented by four genera namely Mastrevirus, Curtovirus, Topocuvirus and Begomovirus2. The genus Begomovirus is the largest and consists of more than 180 species and several unassigned isolates. Begomoviruses is the largest genus of the family of single stranded DNA plant viruses and contains whitefly- transmitted geminiviruses that infect dicotyledonous plant Particle of Gemini viruses are quasi-isometric and they are usually found in pair , hence the name ‘Gemini’3. Symptoms vary from mosaic, yellow leaf, curling, rolling of leaves, thickening of veins, reduction of fruit set, yellow mottling, crinkling, severe stunting, reduced yields and at times death of plants. On infection typical mosaic-like leaf patterns of light and dark green occur4.They are causal agent of some of most devastating plant leaf curls in cotton, pepper and tomatoes, mosaic and yellow mosaic of cassava, pulses and beans. During severe infection,India virus Munbean yellow mosaic virus (MYMIV) and Tomato leaf curl virus (ToLCV) can damage up to 70% and 100% of yield respectively5. The Gemini viruses can be found throughout the subtopic and tropic including Central America and the Caribbean. Year after year in the early 1990s, for example, Gemini viruses destroyed up to 95% of the tomato harvest in the Dominican Republic, and in just the 1991-92 growing season in Florida, they caused $140 million in damage to the tomato crop6. With the virus continuing to spread and other control measures, such as insecticides falling short, plant genetic engineers are struggling to create resistant plants. So far they have had no lasting successes, although they do have promising leads. The viruses have also struck in the Caribbean in the early 1990, the first serious Gemini viruses infections appeared in Florida & Texas in the united state at about the same time7,8,9. As is evident that a begomovirus has immense economic implications and owing to its easy availability it was selected as the candidate for our research project10.
[adsense:468x15:2204050025]
MATERIALS AND METHODS
A range of techniques can be used to detect and identify Gemini viruses based on enzymatic analysis, biochemical analysis, and PCR based analysis11.
Different methods are:
a. Protein based – immunological tests, ELISA, DOT BLAST.
b. Genome based – PCR.
c. Microscopy based – protein immune assay, fluorescence based assay.
Host Plant chosen
The plant species selected for research is Clerodendron inerme (family Verbenaceae) a commonly used hedge plant.C. inerme is valued in landscaping as a groundcover or hedge plant. It has attractive evergreen foliage and fragrant white flowers that form in clusters and are accented by delicate red protruding stamens. Seaside Clerodendron, as its name suggests, grows well along the beach tolerating the salt spray of the ocean and the harsh rays of the sun. It is a versatile plant and can be grown as topiary or as a bonsai. Clerodendron plants growing inand around the university campus at Greater Noida, UP, Indiawhen surveyed was found to be showing yellow mosaic, bright yellow spots along the midrib which coalesce to give mosaic, reduction in leaf size, and stunting of the plant. Soit was imperative to diagnose and investigate the causal agent12.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to PharmaTutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
Methods
Homogenize up to 100 mg wet weight (or up to 200 mg dry weight or lyophilized) plant material. Proceed with buffer LB1 (step 2a) or alternatively with buffer LB2 (step 2b). Transfer the resulting powder (from step 1) to a fresh tube. Add 400µl buffer LB1. Vortex the mixture thoroughly. Add 10µl RNase A solution and mix sample thoroughly. Incubate the suspension at 65°C for 10 minutes. Or Add tube 300µl buffer LB2. Vortex the mixture thoroughly. Add 10µl RNase A solution and mix sample thoroughly. Incubate the suspension at 65°C for 10 minutes. Place a DNAsure® shredder into a fresh collection tube (2ml) and load the lysate onto the Column. Centrifuge for 2 minute at 12000rpm, collect the clear flow-through and discard the DNAsure® shredder mini column. Add 450 µl buffer BBC and mix thoroughly by pipeting up and down (5 times) or by vortexing. Place a DNAsure® plant column into a fresh collection tube (2 ml) and load a maximum of 700µl of the sample. Centrifuge for 1 minute at 1200 rpm and discard the flow through. Add 400µl buffer WB1 to the DNAsure® plant column. Centrifuge for 1 minute at 1200 rpm and discard the flow through. Add 700µl buffer WB2 to the DNAsure® plant column. Centrifuge for 1 minute at 1200 rpm and discard the flow through. Add another 200µl buffer WB2 to the DNAsure® plant column. Centrifuge for 1 minute at 1200 rpm in order to remove wash buffer and dry the silica membrane completely. Place the DNAsure® plant column into a fresh 1.5 ml micro centrifuge tube (not provided). Pipette 50µl buffer PE (70°C) on to the membrane. Incubate the DNAsure® plant column for 5 minute at 70°C. Centrifuge for 1 minute at 1200 rpm to elute the DNA. Repeat this step with another 50µl buffer PE (70°C) and elute into the same tube13.
Primer Designing and PCR Amplification
Primers were manually designed using gene sequences available at NCBI Genbank. Bioedit (7.0.7.1) (Hall, 1999) and CLUSTAL W (Thompson et al., 1994) was used to align the sequences and locate the conserved regions. They were designed with annealing temperatures in the range of 56 – 60oC. The primers were synthesized by Sigma Aldrich. The expected size of the Amplicon was ~750 bp. The names of primer sets, length of primers, approximate amplicon size, annealing temperature are Gemi cp-F (18), Gemi cp-R (21) and 550C, 540C respectively14.
For DNA estimation
Fill the cuvette with water or TE buffer. Zero the spectrophotometer at 260 nm with this blank.DNA from plasmid and genomic preparations is typically at a concentration exceeding 1 micrograms/micro liter. Consequently, DNA is usually diluted before measuring its absorbance. Start by diluting the DNA sample 1 micro liter: 999 micro liters of TE buffer (the dilution can be done directly in the cuvette). Mix the dilution thoroughly.Measure the optical density (OD). Multiply the resulting OD by 50 micrograms/ml. For a 1:1000 dilution, the mass of DNA is equal to micrograms/micro liter. Similarly, the same sample can be measured at 280 nm. A ratio of the OD-260nm/OD-280nm is an indicator of DNA purity. A ratio of 1.8 or higher indicates minimal protein contamination15.
Results and Discussion
736 bp long nucleotide sequences were obtained from the amplified gene sequences (Gemini). The nucleotide sequence showed 99% sequence identity to nucleotides of Coat protein gene of Clerodendron yellow mosaic virus (Indian isolates) (Accession no EF408037.1) and Clerodendrum golden mosaic China virus (Accession no FN645907.1). It also showed 79 to 81% sequence similarity with other Gemini viruses like Cotton leaf curl Burewala virus (Accession no FR750324.1) and Tomato leaf curl Pakistan virus (Accession no DQ116884.1). This nucleotide sequence has a potential to encode a 28.36kDa protein with 244 amino acids residues. The amino acid sequence for the predicted protein shared maximum identity of 98% with capsid protein of Clerodendron yellow mosaic virus amino acid sequences (AAY25168.1) and shared identity of 97 to 98% with other Gemini virus capsid protein sequences. BLAST P analysis of CP indicated that it belonged to protein family pfam00844, a member of the GeminiCP Super-family cl02983.
Accurate detection of begomoviruses is very important due to its wide spread occurrence & its negative effect on health & yield of the host plants. For reliable detection of begomoviruses using ELISA and PCR, knowledge of the serological diversity of a virus is important. Its detection by quicker & reliable methods have been shown earlier. The present study was undertaken to develop more economical & consistent method for detection of Clerodendron yellow mosaic viruswith PCR. Various parameters influence the success of PCR based detection. One of components is preparation of good quality DNA template from plant tissue. In most cases it is achieved by use of commercial DNA extraction kits, which are expensive. Other cheaper method for the extraction of DNA needs to be exploited. In this study the non phenol-chloroform method was found to be quite comparable to commercial DNA extraction. The other important components of PCR based detection are primer specificity & size of amplicon. In this study we could standardize a highly efficient and rapid diagnostic PCR which was based on CP gene of the virus. Coat protein plays an important role in infection and many management strategies uses antisense RNA of CP to develop transgenic against the Gemini virus. Also CP being much conserved due to its importance in disease development is useful for designing primer. Therefore knowledge of sequences of CP would help develop diagnostics as well as management strategies of Gemini virus.
Coat protein

Conclusion
In this project a set of primers was designed from conserved Coat Protein region of Clerodendron yellow mosaic virus which could successfully amplify the CP gene of. Total DNA extraction process from plants was standardized for PCR. A Polymerase Chain Reaction was standardized to amplify the CP gene of Clerodendron yellow mosaic virus. Successful molecular detection of virus infecting Clerodendron inerme by a begomovirus coat protein specific PCR suggested it to be a member of begomovirus. The sequence obtained showed highest similarity with Clerodendron yellow mosaic virusand but had considerable similarity with other begomovirses. Sequencing of the PCR product and sequence analysis by various bioinformatics tool confirmed the pathogen to be Clerodendron yellow mosaic virus, a member of begomovirus group. BLAST P analysis of not sequence obtained indicated that it belonged to protein family pfam00844, a member of the Gemini CP Super-family cl02983.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to PharmaTutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
Acknowledgements:
The authors are thankful to Dr. (Associate Prof) Shahana Majumder, Dept. of Biotechnology, Sharda University, School of Engineering and Technology, Greater Noida, (U.P.),India and Dr. (Prof) S. K. Gupta, Head, Director, Dept of Pharmacognosy, BBS Institute of Pharmaceutical and Allied Sciences, Greater Noida, (U. P.), India for providing the facilities necessary to carry out the survey, study and other activity which are helpful for the research work and to comply my work.
Nucleotide Blast Result
|
Accession |
Description |
Max identity |
|---|---|---|
|
EF408037.1 |
Clerodendron yellow mosaic virus isolate iari, complete genome |
99% |
|
AY950580.1 |
Clerodendron yellow mosaic virus isolate nbpgr coat protein gene, |
99% |
|
FN645907.1 |
Clerodendrum golden mosaic China virus partial AV1 gene for |
99% |
|
FR750324.1 |
Cotton leaf curl Burewala virus complete genome, clone MV18B |
81% |
|
FR750322.1 |
Cotton leaf curl Burewala virus complete genome, clone MV16 |
81% |
|
DQ116884.1 |
Tomato leaf curl Pakistan virus isolate Rahim Yaar Khan 1 clone |
81% |
|
FR837934.1 |
Cotton leaf curl Burewala virus complete genome, clone MV14C |
80% |
|
FR837933.1 |
Cotton leaf curl Burewala virus complete genome, clone MV14A |
80% |
|
FR837932.1 |
Cotton leaf curl Burewala virus complete genome, clone MV13 |
80% |
|
FR750323.1 |
Cotton leaf curl Burewala virus complete genome, clone MV18A |
80% |
|
FR750321.1 |
Cotton leaf curl Burewala virus complete genome, clone MV15 |
80% |
|
FR750318.1 |
Cotton leaf curl Burewala virus complete genome, clone MV12 |
80% |
|
FN645934.1 |
Cotton leaf curl Burewala virus AV1 gene for coat protein, |
80% |
|
FN645932.1 |
Cotton leaf curl Burewala virus segement A, complete sequence, |
80% |
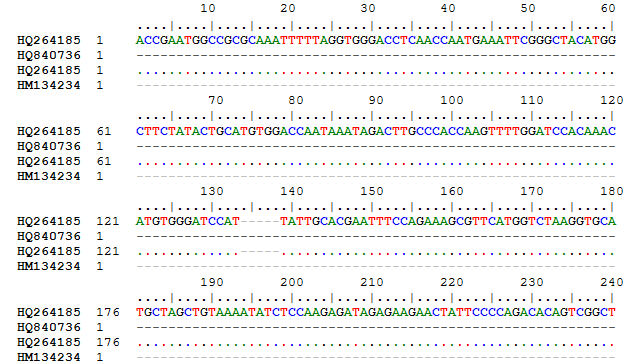
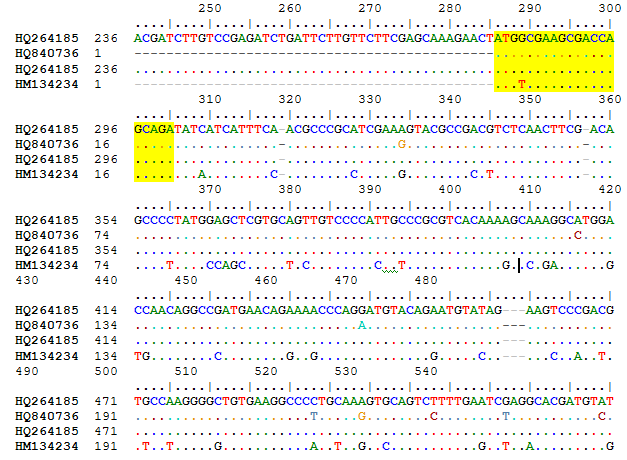
Primer Designing Using Bioinformatics Tool
The highlighted regions were used for designing primers.
REFERENCES
1. Anderson P. K., Cunningham A. A., Patel N. G., Morales F. J., Epstein P. R. and Daszak P., Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evolution, 2004; 19: 535–544.
2. Polston J.E., Hiebert E., Mc Govern R.J., Stansly P.A. and Schuster D.G.: Introduction to tomato yellow leaf curl virus in Florida and implications for the spread of this and other geminiviruses of tomato. Plant Disease, 1993; 77: 1181-11.
3. Autrey L.J.C., Ricaud C., Sullivan S., and Dhayan S., Control of yellow spot disease of sugarcane by aerial application of fungicide, Sugar Azucar, 1983; 78: (7) : 23-25.
4. Arguello-Astorga G.R., Guevara-Gonza´lez R.G., Herrera- Estrella L.R., Rivera Bustamante R.F., Virology, 1994; 203: 90–100.
5. Moffat A., Geminiviruses emerge as serious crop threat, Science, 1999; 286: 1835.
6. Briddon R.W. and Stanley Sub-viral agent associated with plant infecting single stranded viruses virology, 2006; 344:198:210.
7. Luan Y.S., Zhang J., Liu D.M., Li W.L., Virus Genes; 35, 379– 385, 2001; 52: 215–218.
8. Costa A.S, three whitefly-transmitted virus diseases of beans in Sao Paulo, Brazil, FAO Plant Protection Bulletin, 1965; 13: 2-12.
9. Dasgupta I., Malathi V.G. and Mukherjee S. K., Genetic engineering for virus, resistance Curr. Sci., 2003; 84: 341–354.
10. Saunders K., Bedford I.D., Briddon R.W., Markham P.G., Wong S.M. and Stanley J.A. unique virus complex causes Ageratum yellow vein disease; PNAS, 2000; 97: 6890-6895.
11. Xiong Q., Fan S.W., Wu J.X., Zhou X.P., Phytopathology, 2007; 97: 405–411.
12. Faria, J.C., Zerbini E., Familia F.M. Gaminiviridae; Taxonomic, replicacao emovimento, Revisao Annual de patologia de plantas, 2000; 8: 27-57.
13. Fauquet C.M., Briddon R.W., Brown J. K., Moriones E., Stanley J., Zerbini M. and Zhou X., Geminivirus strain demarcation and nomenclature. Arch. Virol., 2008; 153: 783–821.
14. Fauquet C.M., Chatterji, A., M. Padidam, R. N. Beachy, Identification of replication specificity determinants in two strains of tomato leaf curl virus from New Delhi. J. Virol, 1999; 73: 5481-5489.
15. Hanley –Bowdoin L., Settlage S.B., Orozco B.M., Nagar S., Robertson D., Crit. Rev. Plant Sci., 1999; 18: 71–106
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to PharmaTutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE









