 About Authors:
About Authors:
Hardik R. Patel
Industrial Biotechnology from Sardar Patel University,
Gujarat, India.
hardikigbt@gmail.com
ABSTRACT
Cryopreservation and lyophilization of plant germplasm has obvious advantages over in vitro storage in term of space saving and improved phytosanitation. We compared cryopreserved and lyophilized leaf as sources for genomic DNA isolation by CTAB protocol and PVP protocol.Our results showed that cryopreservation of leaf tissue yielded high molecular weight genomic DNA. The DNA was suitable for restriction-enzyme digestion and as a template for polymerase chain reaction (PCR) amplification. While these results rule out cryopreserved tissue as a source for DNA isolation, the ability to freeze-dry, powder, and efficiently store voluminous tissue samples for later use in DNA and protein isolation could be of great benefit to laboratories involved in molecular genetics and molecular biology.
[adsense:336x280:8701650588]
Reference Id: PHARMATUTOR-ART-1563
MATERIALS AND METHODS
Plant Material
Total 4plant species were collectedforDNA isolation from Indroda Botanical Garden, Gandhinagar.
Table 1: Plant species collected from Indrora Botanical Gardenand its GPS location.
|
Plants name |
Family |
Latitude |
Longitude |
|
Acacia Arabica |
Mimosaceae |
N 23° 11.558 |
E 72° 38.939 |
|
Acacia sinuata |
Mimosaceae |
N 23° 11.566 |
E 72° 38.944 |
|
Adenanthera pavonina |
Mimosaceae |
N 23° 11.571 |
E 72° 38.937 |
|
Prosopis spicigera |
Mimosaceae |
N 23° 11.680 |
E 72° 38.836 |
LYOPHILIZATION
Surface sterilization
Leaves were sterilized by surface washing under tap water than distilled water to remove dust and other contaminants.
Sample preparation for Lyophilization
The leaves were dried with blotting paper and middle ribs were removed. They were crushed into liquid nitrogen to make fine powder and approximately 15 g were weighed. After lyophilization, the powder was collected in petridish which were sealed with cling film.
[adsense:468x15:2204050025]
Lyophilization process
Plant samples were lyophilized at vacuum of 0.16 mbar and -50 °C for 72 hrs. Lyophilized powder stored was stored in tightly sealed petridishes at room temperature.
The SuperModulyo freeze dryer used for lyophilization at GSBTM does not facilitate the temperature and pressure regulation. The protocol for operation of this instrument is as follows:
1. The powdered samples were frozen for some time at -20°C freezer.
2. The frozen leaves were evenly spread on the product trays without the delay for the leaves to cool down again.
3. The racks with the samples were placed in the dome and covered with the acrylic lid.
4. It was made sure that all the detachable parts associated with vacuum are well greased so as to minimize the chances of any leakage.
5. The “Fridge” switch was turned on. Within 1 hr, the chamber temperature should be around -40°C to -50°C.
6. The “pump” switch was turned on. The pressure reached up to 10-1 mbar.
7. The samples were lyophilized for 1 whole day.
8. After the run was complete, the “Pump” switch was turned off. The knobs were gradually opened to release the pressure.
9. The samples were placed in a dry, air tight environment as soon as possible and stored out of direct sunlight.
CRYOPRESERVATION
Surface sterilization
Leaves were sterilized by surface washing under tap water than distilled water to remove dust and other contaminants.
Weighing of sample
The leaves were dried with blotting paper and approximately 20 g leaves were weighed without middle ribs.
Sample preparation for cryopreservation
Leaves were crushed into liquid nitrogen to make a fine powder and 3 cryovials were filled up for each plant and were cryopreserved at -196 °C cryopreservation was carried out by placing the cryovials in canisters and immersing them in the Cryocane containing liquid nitrogen. DNA extraction was carried out from cryopreserved sample at intervals of 1, 7 and 15 days.
DNA EXTRACTION
DNA extraction was carried out from cryopreserved, lyophilized and oven dried plant samples using two different protocols which are mentioned below as follows:
A. CTAB protocol [Doyle J and Doyle J, 1987]
1. Water bath was set to 65 ºC and CTAB buffer was put into the water bath.
2. 100 mg of plant material (cryopreserved, lyophilized, oven dried samples) was taken in a motor pestle.
3. Liquid nitrogen was added and the sample was crushed to powder form. (In case of already crushed samples, proceed to next step)
4. 3 ml of 3% CTAB buffer, containing 6% PVP (pH 8) and 6.1 µl of β-mercaptoethanol was added.
5. The solution was incubated at 65ºC for 1 hour.
6. The tube was centrifuged at 12,000 rpm for 5’ at 4 0C by using centrifuge (Eppendorf).
7. The supernatant was collected in fresh tube and 5µl/ml of Proteinase K was added and incubated at 65 °C for 1 hr.
8. Equal volume of P: C: I was added, mixed gently, centrifuged 12000 rpm for 15 mins.
9. Supernatant was collected, equal volume of C: I was added, mixed and centrifuged at 12000 rpm for 15 mins.
10. Step 9 was repeated.
11. 5 µl of 20 mg/ml RNase A or 10µl of 10 mg/ml RNase A was added incubated at 65 °C for 1 hour or 37 °C overnight.
12. 1/10th volume of 3M sodium acetate and equal volume of absolute ethanol was added, incubated for 20 mins at -20°C.
13. The samples were centrifuged at 12000 rpm for 10 mins, supernatant was discarded and the pellet was recovered.
14. The pellet was washed twice with 70% ethanol and centrifuged at 10000 rpm for 10 mins.
15. The pellet was allowed to air dry and dissolved in 200 µl of 1X TE buffer.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
PVP protocol [KIM Cet al., 1997]
1. Approximately 100 mg of sample was ground in a pestle in 5 µl (one drop) of 1% (v/v) 2-mercaptoethanol.
2. After grinding, 300 µl of extraction buffer (250 mM NaCl, 25 mM EDTA, 0.5% SDS, 200 mM Tris–HCl pH 8.0) was added to the homogenate, which was transferred to a 1.5 ml tube and the tube was flicked at the bottom occasionally to keep the extract mixed.
3. The homogenate was incubated at room temperature for 1 hr.
4. Freshly prepared PVP (6% of final volume) and one half volume of 7.5 M ammonium acetate were added separately.
5. The mixture was incubated on ice for 30 min and centrifuged for 10 min in a micro centrifuge (10 000 g at 4 °C).
6. The supernatant was transferred to a fresh tube to which was added 1 volume of isopropanol, and left at -20 °C for 30 min to precipitate the DNA.
7. After centrifugation at 10,000 g for 10 min, the supernatant was discarded and the DNA pellet was air-dried.
8. The DNA pellet was resuspended in 500 µl TE buffer (10 mM Tris–HCl pH 8.0, 0.1 mM EDTA pH 8.0) or distilled water.
9. Two µl of RNase (1 mg/ml) was added to the solution and incubated at 37°C for 15min.
10. One volume chloroform: isoamyl alcohol (24:1) was added and emulsified by inverted shaking to remove both RNase and plant pigments.
11. The above step was repeated once again.
12. After centrifugation (10,000 g at 4 °C) for 5 min, the supernatant was transferred to a fresh tube to which 1 volume of isopropanol was added and left at –20 °C for 10 min.
13. After centrifugation at 10,000 g for 10 min, the pellet was washed with 1 ml 80% ethanol and air-dried.
14. The DNA pellet was re-dissolved in 30 µl TE or distilled water.
AGAROSE GEL ELECTROPHORESIS
Agarose gel electrophoresis is carried out to check the presence of genomic DNA isolated from plant samples.
Method
1. The required amount of agarose was weighed to prepare 0.8% agarose gel in 0.5X TBE buffer. 0.5 µl of ethidium bromide was added.
2. The tank was filled up with 0.5X TBE buffer and immersed the gel in it.
3. 5 µl of sample with 2 µl of bromophenol blue was mixed and the samples were loaded in the respective lanes.
4. The gel was run at 80-100 V for 1 hour.
5. The agarose gel was observed under the UV transilluminator.
6. The image was captured, labelled the required lanes and saved the image.
RESULTS AND DISCUSSION
High quality of DNA, RNA and protein are required for meaningful molecular biology studies. Several extraction methods have recently been developed for DNA, RNA and protein extraction. However, many of these techniques require either fresh tissue or tissue stored and strictly maintained at ultralow low temperatures at -50 ?C to -70 ?C [Saha S et al., 1997]. At this ultra-low temperature, all cellular divisions and metabolic processes are stopped, allowing conservation for a theoretically unlimited period of time [Engelmann, 2004]. Generally, only a limited number of freshly collected tissue samples can be extracted at any given time, while ultralow freezer space for storage of bulky tissue samples is usually at a premium in most laboratories. A solution to this problem is lyophilization of the plant tissues by grinding them as dry powder for efficient storage in limited freezer space [Saha S et al., 1997].
In this study, we tried comparing a protocol for lyophilization, cryopreservation and DNA extraction of four socio-economically and medically important plant species. We also tried to study whether the lyophilized plant leaves were suitable as a source for DNA isolation. A comparative analysis was also performed to find out which among cryopreservation and lyophilization was a better method for storage of plant leaf tissues, by isolation of DNA from the cryopreserved and lyophilized leaves. Similarly, oven drying and lyophilization techniques were also compared for all the plant species studied. Four plant species, Acacia arabica, Acacia sinuata, Adenanthera pavonina, and Prosopis spicigera belonging to Mimosoideae family were selected for the study.
For optimization of DNA extraction method, two protocols were performed one of which was a modified CTAB protocol and the other was PVP protocol. The CTAB protocol gave better results than PVP protocol for DNA isolation from cryopreserved and lyophilized leave samples.
In case of lyophilization, the concentration and the purity of the DNA extracted were in the range of 170-1150 ng/μl and 1.0-2.1 respectively. Bands were observed in case of Acacia arabica, Acacia sinuata, Adenanthera pavonina L. and Prosopis spicigera after storage of the lyophilized plant leaves for up to 15 days [Fig 1]. By PVP absorbance 2.0-2.1 indicates presence of RNA contamination. The residual moisture content post lyophilization was as low as 51.33 % in Acacia arabica and as high as 63.66 % in Prosopis spicigera [Fig. 2].
In case of oven drying, no bands were observed indicating that the plants were sensitive to oven drying. Absorbance at 260/280 was 1.33-1.66 indicates readings from very dilute samples, phenol and other contaminants, acidic solutions,or the nucleotide composition of the bases present in DNA.
In case of cryopreservation, the concentration and the purity of the DNA extracted was in the range of 17-1100 ng/µl and 1-1.8 respectively. Bands were observed in cryopreserved leaf samples of Acacia arabica, Acacia sinuata, Adenanthera pavonina L., and Prosopis spicigera stored for up to 15 days [Fig 4].
After storage of some plant species post lyophilization and cryopreservation, concentration and purity of the DNA extracted on nanospectrophotometer but no bands were observed after agarose gel electrophoresis. Many factors affect the preservation of DNA, including the type of plant, the chemical and physical environment in which that plant is stored, and the duration of storage and the preservation method. However, the interactions of these factors and their resultant effects on DNA preservation are difficult to predict [Dawson M et al., 1998].
The purity of the DNA extracted from both cryopreserved and lyophilized leaves was in the range of 1-1.8. According to Reddy J, a ratio less than 1.8 indicates the probable presence of proteins and/or UV absorbers. A ratio higher than 2.0 indicates that the sample may be contaminated with chloroform or phenol.
In the comparative study of lyophilization and cryopreservation of Acacia arabica, Acacia sinuata, Adenanthera pavonina, and Prosopis spicigera both techniques gave results but cryopreservation of samples give better results than lyophilization. CTAB protocol gives high quality and quantity of genomic DNA compared to PVP protocol in case of all plant species. [Fig 3, 4]
In case of lyophilization Acacia arabica, Acacia sinuata gives high quality of DNA and in case of cryopreservation Acacia arabica and Adenanthera pavonina gives high quality of DNA.
Similar results were obtained by other authors who used optimizing protocols for extracting DNA from legume by using the CTAB method for ensuring purity and quantity. Chiari et al. (2009) obtained 13.30-15.84 μg DNA from 300 mg lead tissue of Stylosanthes guianensis, with purity ratios from 1.40 to 1.50 nm. Ginwal and Mawrya (2009) successfully optimized the CTAB protocol for extracting DNA from Dalbergia sissoo and obtained significant DNA amounts with purity ratios of 1.71-1.90 nm.
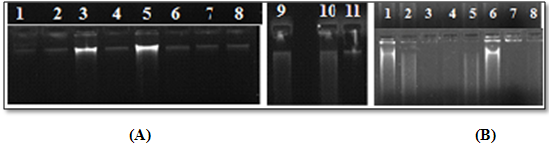
Figure 1: Agarose gel electrophoresis of genomic DNA using (A)CTAB and (B) PVP method, of leaf samples stored for 15 days post lyophilization:
(A) CTAB method left to right lane 1, 2:Acacia sinuata; lane 3, 4, 5: Acacia arabica; lane6, 7, 8:Adenanthera pavonina;lane 9, 10, 11: Prosopis spicigera.
(B) PVP method from left to right lane 1, 2: Acacia arabica;lane 1, 2:Acacia sinuata; lane 1, 2:Adenanthera pavonina; lane 1, 2: Prosopis spicigera.
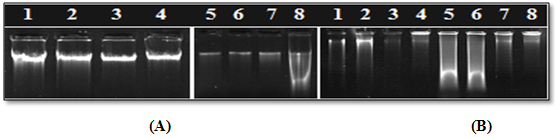
Figure 2: Agarose gel electrophoresis of extracted genomic DNA using (A) CTAB and (B)PVP method, for leaf samples stored for 15 days post cryopreservation:
(A) CTAB method from left to right lane 1, 2: Acacia arabica;lane 3, 4:Acacia sinuata;lane 5, 6:Adenanthera pavonina;lane 7, 8:Prosopis spicigera
(B) PVP method from left to rightlane 1, 2: Acacia arabica;lane 3, 4: Prosopis spicigera; lane 5, 6:Acacia sinuata;lane 7, 8:Adenanthera pavonina
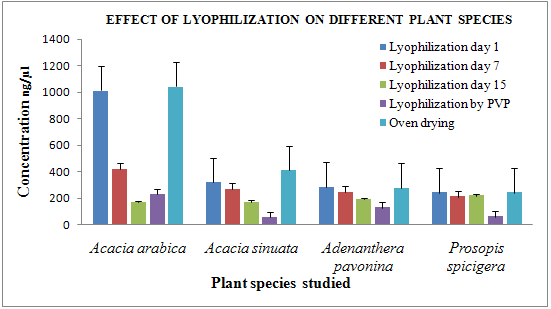
Figure 3 Effect of lyophilization on different plant species
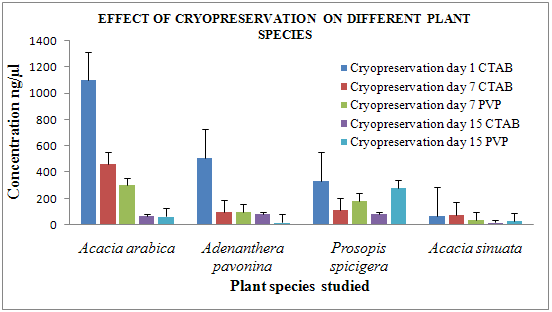
Figure 4 Effect of cryopreservation on different plant species
CONCLUSIONS
· Since decades, lyophilization and cryopreservation are the techniques which have been used for the long term preservation of plant germplasm
· CTAB method gave better results as compared to PVP method in case of all the plant species studied. The concentration and purity (A260/A280) of the DNA extracted using CTAB protocol ranged from 57 - 1100 ng/µl and 0.716 – 2.010 respectively. A purity ratio less than 1.8 indicates the probable presence of proteins and/or UV absorbers.
· The residual moisture content post lyophilization ranged from 50.66 % in Acacia arabica 65 % Acacia sinuata.
· Many factors affect the preservation of DNA, including the type of plant, the chemical and physical environment in which that plant is stored, and the duration of storage and the preservation method. However, the interactions of these factors and their resultant effects on DNA preservation are difficult to predict and the protocol for DNA extraction needs to be further optimized.
· From this, it may be concluded that cryopreservation seems to be an appropriate storage method for the above mentioned plant species as compared to lyophilization
ACKNOWLEDGMENTS
A word of appreciation to Dr. Madhavi Joshi, Dr. Snehal Bagatharia, Dr. Rohan Pandya, Miss. Bala Iyer,Mrs. Arti Bambhaniya for her help in DNA extractions and for their helpful suggestions on the manuscript. Finally special thanks to my family members for their support.
REFERENCES
• Doyle JJ and Doyle JL., A rapid DNA isolation procedure for small quantities of fresh leaf tissue; Phytochemical Bulletin 1987, 19: 11-15
• Doyle JJ, Doyle JL., A rapid DNA isolation procedure for small quantities of fresh leaf tissue; Phytochemical Bulletin 1987, 19:11-15.
• Kim CS, Lee, CH, Shin JS, Chung YS, Hyung NI., A simple and rapid method for isolation of high quality genomic DNA from fruit trees and conifers using PVP; Nucleic Acids Research 1997, 25(5):1085-1086.
• Saha S., Challahan FE, Dollar DA, Creech JB., Effect of Lyophilization of Cotton Tissue on Quality of Extractable DNA, RNA, and Protein; The Journal of Cotton Science 1997, 1:10-14
• Engelmann F., In Vitro Cell Dev Biol Plant 40; 2004; 427-433.
• Saha S., Challahan FE, Dollar DA, Creech JB., Effect of Lyophilization of Cotton Tissue on Quality of Extractable DNA, RNA, and Protein; The Journal of Cotton Science 1997, 1:10-14
• Chiari L, Valle JVR and Resende RMS (2009)., Comparação de três métodos de extração de DNA genômico para análises moleculares em Stylosanthes guianensis; Circular Técnica 36: 1-6.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE









