 About Authors:
About Authors:
Dhanya V C
Kasturba medical college, Manipal University
Indian Veterinary Research Institute
dhanya285@gmail.com
INTRODUCTION:
A biosensor is a device that uses specific biochemical reactions mediated by isolated enzymes, immunosystems, tissues, organelles or whole cells to detect chemical compounds usually by electrical, thermal or optical signals (1). Or in other words, Sensor that integrates a biological element with a physiochemical transducer to produce an electronic signal proportional to a single analyte which is then conveyed to a detector.
[adsense:336x280:8701650588]
There are 4 aspects to making a measurement-
• sampling
• sample processing
• analysis
• Data processing.
Biosensors combine all 4 of these aspects.
- Sampling occurs when wherever the sensor is placed
- Sample processing is typically eliminated because the sensor contains all the necessary components to provide specificity and sensitivity.
- Analysis is accomplished by binding of the analyte to the sensor
- Data processing is performed on the signal generated from the sensor.
Reference Id: PHARMATUTOR-ART-1610
So needless to say, a properly designed nano-scale biosensors have many advantages including: i) high sensitivity due to comparable size of sensor element (nm to µm) and analyte/target species (0.2 to 2 µm) and high surface area to volume ratio, ii) low cost of materials and the reagents, iii) shorter assay time, iv) portability and use as point-of-use or point-of-care devices.
In the proposed study, we are interested in developing a biosensor for HIV diagnosis. Even though we have many specific serological tests available for HIV diagnosis, none of the serological test is useful to detect the infection in all the stages.Challenges of HIV Testing includes: Sensitivity- Early diagnostic (window period), specificity- cross reactivity, easy to perform, low cost and detection of HIV-1 and HIV-2 and discrimination between the two viruses. None of the currently available tests fulfill all these requirements. Hence there is a need to perform a combination of HIV tests for screening and confirmation.
Currently, HIV diagnosis is accomplished by following ERS strategy (ELISA/ Rapid test/ Simple test) (2). There are strategy 1, 2 and 3; strategy one includes testing with a single ERS test. Strategy 2 includes supplemental ERS testing of all samples reactive in an initial ERS test and strategy 3 includes supplemental ERS testing of all samples reactive in each of two sequential ERS tests.
In many laboratories testing for antibodies to human immunodeficiency virus HIV) (both HIV-1 and HIV-2) currently implies carrying out Western blot (WB) testing of sera found reactive in initial screening by simpler tests, such as enzyme-linked immunosorbent assays (ELISA). WB testing is relatively expensive and technically demanding.
PCR can detect HIV in all stages. But it is complex and costly and indicated only when other methods cannot give a definitive result.
[adsense:468x15:2204050025]
So if we can develop a biosensor with 100% sensitivity and specificity which can give accurate results at all stages it will be of a significant achievement with regard to saving cost, time, labor and resources.
It is estimated that, laboratory HIV testing accounts for approximately 65% of the expenditure on materials and supplies of National AIDS Programme.
Therefore this research is carried out with an aim to develop a diagnostic biosensor for use in settings where the resources are limited thereby supporting the global prevention and control of HIV infection.
The principle behind the idea is to utilize the basic concept of the HIV pathogenesis. That is, all HIV viruses use CD4 molecule, which is expressed on macrophages and T-lymphocytes, as a receptor onto which HIV envelop glycoprotein 120 (gp 120) binds. A second co-receptor (CCR5 or CXCR4) in addition to CD4 is necessary for the effective binding of HIV.
The mechanism is diagrammatically represented as follows:
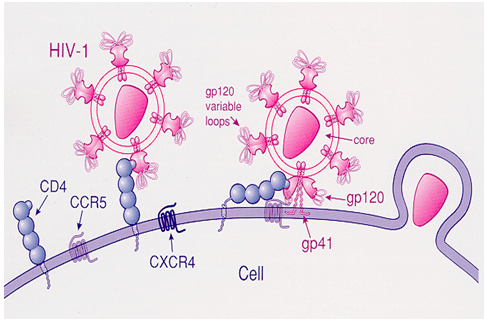
A biosensor with all possible viral receptor, antigens and antibodies attached to the sensing device possibly detect HIV at all stages. This is what we are trying to accomplish. To the best of our knowledge such a detecting system for HIV diagnosis is not currently available.
And it is based on the concept of taking ideas from nature. Cell membrane may be considered a completely self-contained biosensing system wherein molecular recognition is directly linked to signal transduction (3). The synthetic membranes are organized supramolecular structures that resemble natural cell surfaces at the interfacial region.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
If it is possible to couple a chromophoric conjugated polymer at its interior, the latter part serves as an optical “transducer” of molecular recognition events occurring at the interface. Signaling occurs by a simple color change of the chromophoric unit. We are also aiming tocheck whether we can develop a signal similar to the signal transduction pathways taking place in our body. Main purpose of our study is to obtain new facts about the generation of immune signals in an artificial liposome when a receptor-ligand interact with each other, that can be used as a principle for developing a diagnostic biosensor for Human Immuno Deficiency Virus or which can also be extended to any other disease markers for our future research works.
So the proposed study is undertaken with the following aims and objectives.
AIM
To develop a diagnostic biosensor for detecting Human Immuno Deficiency virus at all stages of the infection
OBJECTIVES
- To study the analyte - bioreceptor interaction on an artificial liposome.
- To study the immune signal transduction mechanism generated inside the liposomes by analyte-bioreceptor interaction.
· To develop a diagnostic biosensor based on generation of immune signals in the artificial liposome using HIV as a model.
DESIGN OF THE EXPERIMENT
Steps included are:
STEP 1:
Liposome synthesis:
This can be accomplished by following ways:
1. Empty liposomes are commercially available which are actually meant for drug delivery experiments. Details of such a product are given below. Freeze-dried ready-to-use liposome powder, called Empty Liposome (COATSOME® EL) (4), is composed of several kinds of phospholipids and electrolytes. When a drug solution is poured into a vial and gently shaken, the drug (we have to see whether we can use antigens or antibodies instead of drugs) is easily encapsulated in the liposomes. Liposomes with surface modified by PEG derivatives are available (Doxil® / Caelyx™ ;). PEG-conjugated phospholipids are used approximately 6 mol % in all the lipids consisting of the liposome, and the liposome diameter ranges from 100 - 200 nm. Furthermore, polyglycerin (PG), an oligomer of glycerin, can also show prolonged retention in circulation when it is used as the aqueous high polymer instead of PEG.
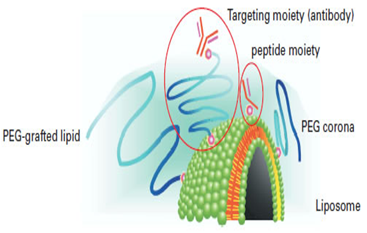
Immunoliposomes with antigen or antibody encapsulated are also available. Two types of approach were considered; the antibody is conjugated directly to phospholipid, or else to an end of the PEG chain. Experimental results indicated that binding to an end of the PEG chain is essential to preserve antigen recognition capacity. Antibody-conjugated liposome is also called pendant-type immunoliposome because of its shape. Pendant-type immunoliposome is expected to play an important role in active targeting, since it has long retention due to PEG and antigen recognition capacity thanks to antibody conjugation.
An immunoliposome using Fab' fragment that lacks Fc region is now available. Longer retention is achieved by Fab'-PEG-liposome. From all these results, Fab' modification on the liposomal surface is considered to be effective in targeting using immunoliposome (5). Use of functionalized phospholipids enables researchers to attach various peptides or other biologics to the surfaces of lipid emulsions or liposomes.
2. Second option is to synthesize a liposome using an instrument named- LIPONIZER® for Liposome production. (LIPONIZER® is a registered trademark of Nomura Micro Science Co., Ltd.)

3. Third option is to synthesize a lipid bilayer with the automated set up as follows:
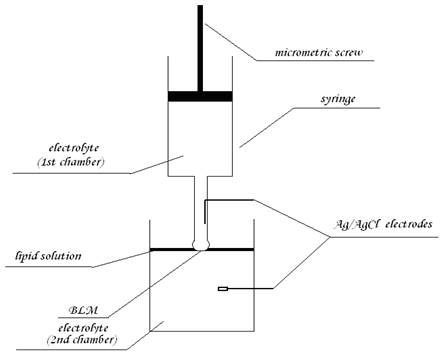
Automated setup for formation of planar lipid bilayers:
The planar lipid bilayers can be made by deposition of a hanging drop of electrolyte on the surface of a thin layer of phospholipid solution swimming on the electrolyte solution (6). Due to gravity, the drop passes through the lipid solution, and a lipid bilayer is formed between these (the drop and the solution) aqueous phases. The membrane can be formed simply by switching on the motor connected with the syringe, and both lipid and lower aqueous solutions can be easily modified or replaced (by peristaltic pump or direct additions), provide a simple way for automated membrane preparation.
Liposomes can be prepared by a variety of methods like ethanol injection method, sonication of water dispersion of lipids by ultrasound, by detergent analysis, by extrusion method, etc. Liposomes are made on demand from Companies like - 1.Encapsula NanoSciences and 2. Avanti lipids
So once liposomes are ready, next step is to bind our bioreceptor (antigen/antibody/protein) on to the liposomes. To increase the affinity of receptor–ligand interaction we can use high affinity intermolecular binding proteins such as biotin-avidin, protein A or G (7). Most commonly and successfully employed one is biotin-avidin interaction.
STEP 2:
Labeling of liposomes with biotin:
Many companies provide biotin labeling kits. One such company product- Lightning-LinkTM conjugation kit allows biotinylation simply by adding a solution of the protein to be labeled to a lyophilized mixture containing a proprietary activated biotin ligand. Upon dissolution of Lightning-LinkTM mixture (8) with a solution of the antibody (or other biomolecule to be labeled so also liposome) proprietary chemicals in the mixture become activated. This results in coupling of the antibody (liposomes) to the biotin, at near-neutral pH.
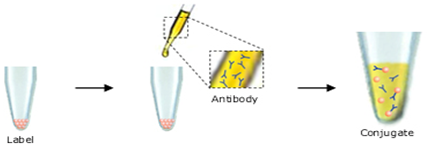
So, once biotin labeled liposomes is available, next step is to attach the biorecptor on to the liposomal surface. For this, bioreceptor has to be labeled with avidin. Thus biotin-avidin high affinity interaction can favor their attachment. Another point to consider is whether we need a solid support on to which liposomes are attached. So that, this solid support can acts as a connecting link between liposomes and the transducer surface.
Most widely used solid support uses covalent immobilization procedures utilizing silanes. Stable lipid bilayers can also be successfully created on agar-agar surfaces or agarose or on the surface of a conducting polymer. Agarose will be an easy option because of its easy availability.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
STEP 3:
Making of a solid support- agarose:
A product named Avidin-agarose is commercially available (company named Uptima). (Avidin immobilized onto an agarose gel).
So the idea is to develop a liposome with biotin at its entire surface. (Imaginary idea- that is; consider liposome as a sphere with two hemisphere, say northern and southern or upside or downside) If we can entrap one hemisphere of the liposome to the agarose layer which is avidin immobilized in it (using the biotin-avidin interaction which can favor their entrapment) and to the other hemisphere avidin labeling receptors of our interest has to be attached. (Refer step 4).
In short, we have liposomes with biotin labeled on one of its surface. We also have antibodies with avidin labeled. We have an avidin immobilized agarose gel. So biotin-avidin interaction favors interaction between receptors to liposomes as well as liposomes to agarose.
Once biotin labeled liposomes are attached to avidin immobilized agarose support, next step is to label the bioreceptor with avidin. Another method is to use agarose micro beads in an array of bottomless wells etched in silicon or perforated in stainless steel.
STEP 4:
Availability of bioreceptors:
Bioreceptors
1. P 24 ANTIGEN
2. ANTI P 24 ( ANTIBODY )
3. CD 4 T CELL
4. CCR5
5. CXCR4
6. ANTI HIV GP 120 IGG
7. HIV 2 GP 39 ANTIBODY
Some of the products available in the market and their details are given below:
1.P24 PROTEIN(HIV-1/HXBc2)
CATALOG NUMBER: IV-017-005P, 50 μg
2. Anti-p24(HIV-1/Clade B)
CATALOG NUMBER: IV-017-0100, 100 μg (E enzyme)
Reacts with HIV-1 p24 and Gag (Clade B) proteins. Cross-reactivity to p24
And Gag from other glades not tested.
3. HIV-Igp120 antibody (pAb) & (mAb)
Rabbit Anti-HIV-1 (gp120), Catalog Number PK-AB913-210
Specifically recognizes HIV-1 gp120.
4. Human CD4 Subset Mini Column Kit
Catalog Number: HCD43
R&D Systems, Inc.
1-800-343-7475
Or
PlusCellect™
Human CD4 Kit
Catalog Number PLS379
For the isolation of CD4 expressing cells
5. DC-SIGN (ED) antibody (pAb)
Rabbit Anti-Human DC-SIGN (Novel HIV Binding Protein), cat no. PK-AB718-2349
6. CCR5 (NT) antibody (pAb)
Rabbit Anti-Human CC-Chemokine receptor 5 (HIV & Chemokine Receptor), Catalog Number PK-AB718-1112
7. CXCR4 (NT) antibody (pAb)
Rabbit Anti-Human/Mouse CX3C-Chemokine Receptor 4 (HIV Receptor), Catalog Number PK-AB718-1009
8.Anti-gp120/160 (HIV-1/Clade B)
CATALOG NUMBER: IV-002-0100, 100 μg
9. Anti-Gag (Pan) (HIV-1)
CATALOG NUMBER: IV-007A-0100, 100 μg
10. Anti-Nef (Clade B)
CATALOG NUMBER: IV-014-0100, 100 μg
11.HIV-Igp41 antibody (mAb)
Mouse Anti-HIV-I gp41, Catalog Number PK-AB913-159
12. HIV-IIgp39 antibody
Rabbit Anti-Human HIV-II gp39, Catalog Number PK-AB913-161
STEP 5:
Labeling of bioreceptors with avidin:
This is achieved in a similar manner described above for biotin labeling. Lightning-LinkTM conjugation kits are available for avidin labeling. The kit allows avidin conjugations to be set up in seconds, simply by adding a solution of the protein (so also our antigen/antibody/bioreceptor) to be labeled to the lyophilized mixture containing a proprietary activated avidin ligand.
Upon dissolution of Lightning-LinkTM mixture with a solution of the antibody (or other biomolecule to be labeled) proprietary chemicals in the mixture become activated. This results in coupling of the antibody to the avidin, in a gentle and controlled process at near-neutral pH.
Many kits are commercially available for labeling proteins or any other biomolecules with avidin or biotin or enzymes or fluorescent dyes. There are many other means of entrapping bioreceptor onto the liposomes.
Some of them include- Methods involving the use of organic solvent, methods involving the use of mechanical means, methods involving the use of detergents, direct incorporation of the proteins into the preformed liposomes, binding of proteins and peptides to liposomes via amino acid groups, binding of proteins and peptides to liposomes via sulfhydryl groups etc. Most convenient and widely used method is biotin-avidin labeling.
If Protein A is used as an intermediate molecule, there is an added advantage. That is, Protein A (protein isolated from the cell wall of Staphylococcus aureus) which interacts specifically with the Fc fragment of IgG (9). After adsorption of Protein A on the surface, the antibody can be simply added. Under these conditions, IgG is correctly oriented with the Fab’ fragment and is accessible for binding to the specific antigen. (e.g.; consider P 24 antigen binding to anti P 24 antibody. So, antibody will bind with the entrapped Protein A with its Fc portion and Fab will be free to combine with antigen. But this is applicable only to our antigen analytes. But we are also interested in antibodies against HIV and also the whole virus with its glycoprotein spikes)
Another point to be considered is what kind of a signal the interaction may generate. That is what we actually need to demonstrate. But for biosensors employing optical signaling principles, for example- let us consider the principle of FRET or use of gold nano particles. Then use of chromophores to make use in FRET or the use of colored gold nano particles has to be considered at the stage of developing liposomes itself.
STEP 6:
Entrapping fluorescent proteins or gold nano particles inside the liposomes for optical signal generation (10, 11)
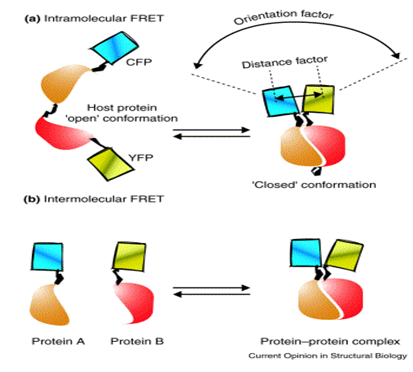
Use of FRET microscopy technique: The combination of enhanced cyan (CFP) and yellow (YFP) fluorescent proteins has proven to be most effective in many FRET studies.(Company supplying YFP: Biovision). Recombinant YFP can be conjugated to other proteins. (So also to our antigen or antibody) It is claimed that YFP can be fused with proteins of interest without interfering significantly with their assembly and function. Biovision also supplies, Recombinant Cyan Fluorescent Protein (CFP).Certain companies like Genova- their antibody processing wing can provide antibody labeling according to customers instructions. So we have to enquire whether they can provide us with our antigen or antibody of interest with cyan fluorescent protein or YFP labels. Protein labeling kits are commercially available (lightning link-a company, this is similar to biotin or avidin labeling kits) which can be used to label these fluorescent proteins on to our protein of interest- antigen/ antibody/ receptor.
Intramolecular FRET can occur when both the donor and acceptor (here donor and acceptor may be our analyte and receptor respectively) chromophores are on the same host molecule, which undergoes a transition, for example, between ‘open’ and ‘closed’ conformations. So before the analyte binds to the receptor the receptor may be in an open conformation, but once the analyte binds to it, it may undergo certain conformation say – a closed one.
The amount of FRET transferred strongly depends on the relative orientation and distance between the donor and acceptor chromophores: the parallel orientation and the shorter distance (<100 Å) generally yield larger FRET.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
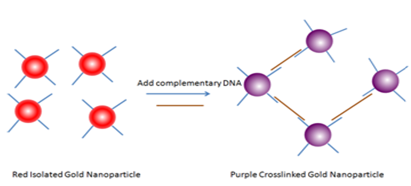
Use of gold nano particles:
Several bio-recognition events have been detected by utilizing the distance dependent color change in gold nanoparticles. (Few examples are a) Streptavidin-Biotin; b) Antibody-antigen; c) Lectin-sugar; d) Aptamer-analytes; e) protein-protein interactions; etc.) Turning the events around-any biological event that can reverse the aggregation can also be detected using gold nanoparticles.
An example is depicted below (picture). Here, complementary DNA is the analyte. We have to see whether our bioreceptor – analyte interaction can bring any colour change if a colored gold nano paricle is attached to the inner layer of liposomes.
FLOW CHART OF THE PROPOSED WORK PLAN
Buy liposomes
↓
Label liposomes with biotin using biotin labeling kits or buy commercially available biotinylated liposomes
↓
Buy antigens, antibodies, receptors & co receptors of HIV
↓
Label these proteins with avidin or buy commercially available avidin labeled proteins of our interest
↓
Attach avidin labeled receptors onto biotin labeled liposomes on one of its sides
↓
Buy avidin labeled agarose
↓
Attach biotin labeled liposomes to avidin labeled agarose on its other side which is free of receptors
↓
To use FRET microscopic principle, label analyte with yellow fluorescent proteins and receptor with cyan fluorescent proteins using protein labeling kits or buy commercially available fluorescent labeled bioreceptor of our interest
↓
Study receptor analyte interaction
↓
Attach agarose onto transducer surface
↓
Convert the optical signal into electrical signal with the help of a transducer
↓
Measure the signal
↓
Interpret the data
References:
1. IUPAC Compendium of Chemical Terminology, 2nd Edn, 1997. Available from <iupac.org/goldbook/B00663.pdf.
2. van der Groen G et al. Simplified and less expensive confirmatory HIV testing. Bulletin of the World Health Organization, 1991, 69: 747-752.
3. Charych; Deborah, Polymeric assemblies for sensitive colorimetric assays, patent filed July 28, 1997. Available from < techportal.eere.energy.gov/patent.do/ID=10873>
4. Empty Liposomes <COATSOME® EL Series. Available from < phospholipid.jp/phospholipid_2-6.html>
5. Kazuo Maruyama. PEG-Immunoliposome. Bioscience Reports, Vol. 22, No. 2, April 2002
6. The National Nanotechnology Infrastructure Network Research Experience for Undergraduates Program, 2009. Available from: <nnin.org/sites/default/files/files/2009reura/2009NNINreuRA.pdf>.
7. James D. Hirsch, et. al. Easily reversible desthiobiotin binding to streptavidin, avidin,and other biotin-binding proteins: uses for protein labeling,detection, and isolation. Analytical Biochemistry 308 (2002) 343–357.
8. Lightning-LinkTM by Innova Biosciences, Available from biomol.de/details/IN/Lightning-Link_biomol.pdf
9. Emmanuel Mongodin, et. al. Cell Wall-associated Protein A as a Tool for Immunolocalization of Staphylococcus aureus in Infected Human Airway Epithelium, J Histochem Cytochem April 2000 vol. 48 no. 4 523-533
10. Sarah J. Leung, et. al. Light-Activated Content Release from Liposomes, Theranostics 2012; 2(10):1020-1036.
11. Weibo Cai, et. al. Applications of gold nanoparticles in cancer nanotechnology. Nanotechnology, Science and Applications 2008:1 17–32
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE









