{ DOWNLOAD AS PDF }
 About Author
About Author
Nirav Soni
Department of Quality Assurance,
A-One Pharmacy College,
Anasan, Ahmedabad, Gujarat, India
nirav_sonic@yahoo.com
ABSTRACT
Isolation of Nucleic acid easily by Solid Phase Extraction (SPE) and this approach, using commercially available Extraction of nucleic acid column-based kits, requires no toxic chemicals and is a rapid and consistent method for concomitant protein extraction. It is a modern technique useful for separation of Nucleic acid which is most reliable, less time consuming and separation of impurities and continuity of reactive products using listed techniques like column-based nucleic acid purification, Nucleic acid methods& ethanol precipitation, DNA separation by silica adsorption. Buffer choice is significant to completely solubilized all proteins in the sample. This technique provides a simple and effective way to analyze protein and nucleic acids simultaneously from the same sample not affecting yield and quality.
[adsense:336x280:8701650588]
REFERENCE ID: PHARMATUTOR-ART-2414
|
PharmaTutor (Print-ISSN: 2394 - 6679; e-ISSN: 2347 - 7881) Volume 4, Issue 6 Received On: 16/01/2016; Accepted On: 03/02/2016; Published On: 01/06/2016 How to cite this article: Soni N; Extraction and Purification of Nucleic Acid using CBNP & PCIA Technique; PharmaTutor; 2016; 4(6); 20-22 |
Column-Based Nucleic Acid Purification (CBNP Method):
CBNP is a Solid Phase Extraction (SPE) method to promptly purify nucleic acids. This method relies on the fact that the nucleic acid may bind based on adsorption to the solid phase using Silica or other which is depend on the pH & the salt content of the buffer which may be a Phosphate buffer or Tris-EDTA (TE) buffer ( used in DNA microarray experiments due to the reactive amines).
Therefore, stages are below (Lyse →Bind→ Wash→ Elute) [1,2] :-
↓Lyse
“lysis”Procedure i.e. Breakdown . The sample cells are broken open with a
↓ Bind
A buffer solution + sample solution along with IPA (Iso Propyl Alcohol) or ethanol .This forms the “binding” solution. The binding solution directly transferred into a spin column and the column is lay down in a centrifuge. The centrifuge forces the binding solution passing through a silica gel membrane that is inside the spin column. If the pH & salt concentration of the binding solution are optimal, the nucleic acid will bind to the silica gel membrane as the solution passes though.
↓Wash
The flow-through is detached & added wash buffer into the column. The column is put in a centrifuge again, forcing the membrane via the wash buffer. This removes any type of present impurities from the membrane, leaving only the nucleic acid bound to the silica gel.
↓Elute
Removed wash buffer and an elution buffer or simply water is added to the column. The column is put into a centrifuge again, forcing the membrane during the elution buffer. The elution buffer which removes the nucleic acid from the membrane and the nucleic acid where it is collected from the bottom of the column.
Procedure :-
1. The sample is added to the column and the nucleic acid binds due to the lower pH (relative to the silanol groups on the column) and salt concentration of the binding solution, which may contain buffer, a denaturing agent (such as guanidine hydrochloride), Triton X-100, isopropanol and a pH indicator [3].
2. The column is then washed (5 mM KPO4 pH 8.0 or similar, 80% EtOH)
3. The column can be eluted with buffer or simply water.
Even prior to the major techniques employed today it was known that in the presence of chaotropic agents, such as sodium iodide or sodium perchlorate[4]. DNA binds to silica, glass particles or to unicellular algae called diatoms which shield their cell walls with silica. This property was used to purify nucleic acid using glass powder or silica beads under alkaline conditions. This was later improved used guanidinium thiocyanate or guanidinium hydrochloride as the chaotropic agent .The use of beads was later changed to minicolumns

[ Fig.1 Silica in a spin column with water and with DNA sample inchaotropic buffer ]
[adsense:468x15:2204050025]
Mechanism of Work :-
This method containing centrifugation process which is used for formation of biphasic mixture or separation of two layer which containing aqueous phase and organic phase such as chloroform, ethanol, alcohol, phenol and a chaotropic denaturing solution i.e. guanidinium thiocyanate resulting in an upper aqueous phase and a lower organic phase mainly chloroform. Nucleic acid partitions in the aqueous phase, while protein partitions in organic phase.[5]
In a final step, RNA is improved from the aqueous phase by precipitation with 2-propanol or ethanol. DNA will be placed in the aqueous phase in the absence of guanidinium thiocyanate and hence, the technique can be used for DNA purification alone.
Guanidinium thiocyanate which denatures proteins including RNases and separates rRNA from ribosomes, while phenol, isopropanol and water are solvents with poor solubility. In the +ve of chloroform or BCP - bromochloropropane, these solvents separate fully into two phases that are known by their color: a clear, upper aqueous phase which containing the nucleic acids and a bright pink lower phase which containing the proteins dissolved in phenol & the lipids itself dissolved in chloroform. Other denaturing chemicals such as 2-mercaptoethanol and sarcosine may also be used. The major downside is that phenol and chloroform both are toxic and not convenient materials and the extraction is regularly laborious, so in recent years many companies now offer another ways to separate DNA.[6,7]
Reagents [8,9,10] :-
Phenol: The phenol used for biochemistry comes as a water-saturated solution with Tris buffer, as a Tris-buffered 50% phenol, 50% chloroform solution, or as a Tris-buffered 50% phenol, 48% chloroform, 2% isoamyl alcohol solution it sometimes called "25:24:1". Phenol is obviously somewhat water-soluble, and gives a blurry interface, which is sharpened by the resulting of + ve chloroform and the isoamyl alcohol which reduces foaming. nearly all solutions also have an antioxidant, as oxidized phenol damages the nucleic acids. For RNA purification, the pH is kept around pH 4, which retains RNA in the aqueous phase preferentially. For DNA purification, the pH is usually near 7, at which point all nucleic acids are found in the aqueous phase.
Chloroform: It is stabilized with small quantities of ethanol or amylene, because exposure of pure chloroform to oxygen and UV light which generates phosgene gas. Some chloroform solutions come as pre-made a 96% chloroform, 4% isoamyl alcohol mixtures that can be mixed with an equal volume of phenol to acquire the 25:24:1 solution.
Phenol–Chloroform Extraction (PC or PCIA) :
Phenol–Chloroform Extraction (abbreviated PC or PCIA, see reagents below) is a Liquid–Liquid Extraction (LLE) technique widely used in the biochemistry & molecular biology for isolation of DNA, RNA and protein. To take a equal volumes of a Phenol: Chloroform (P:C) mixture and an aqueous sample are mixed together resulting to form a biphasic mixture. This method may take longer than a column-based system such as the silica-based purification,
Advantages:-Higher purity and high recovery of RNA: an RNA column is usually not suitable for purification of short nucleotides which is <200. RNA species, such as siRNA, miRNA, gRNA and tRNA. Column methods also shear large DNA fragments, which vary with creates a problem depending on downstream applications.
Isoamyl alcohol: It may reduce foaming & make sure deactivation of “RNase”
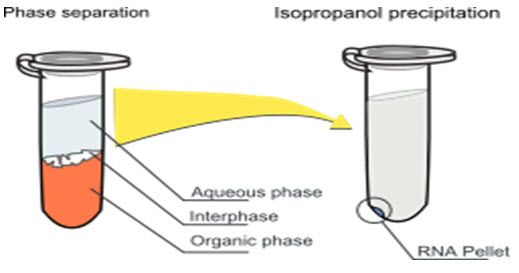
[ Fig.2 Phase Separation and isopropanol Precipitation Method ]
REFERENCES
1. Matson, Robert S;Microarray Methods and Protocols. Boca Raton, Florida: CRC. 2008; 27–29.
2. Kumar; Anil.Genetic Engineering. New York: Nova Science Publishers; 2006;101–102.
3. Marko MA, Chipperfield R, Birnboim HC. A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem.1982 Apr; 121(2):382-7.
4. Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990 Mar; 28(3):495-503.
5. Perry RP, La Torre J, Kelley DE, Greenberg JR. On the lability of poly(A) sequences during extraction of messenger RNA from polyribosomes. Biochim. Biophys. Acta 262: 220–226
6. Chomczynski, P. & Sacchi, N; "Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction". Anal. Biochem.; 162: 156–159.
7. Chomczynski, P. & Sacchi, N."Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on".;Nature Prot. 1 (2): 581–585.
8. Brawerman G, Mendecki J, Lee SY. A procedure for the isolation of mammalian messenger ribonucleic acid. Biochemistry 11: 637–641.
9. Dellaporta, S.L., Wood, J. and Hicks, J.B. A plant DNA mini preparation: Version II. Plant Molecular Biology Reporter 1: 19-21.
10. Saghai-Maroof, M.A., Soliman, K.M., Jorgensen, R.A. and Allard, R.W. 1. Ribosomal DNA spacer length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc. Natl. Acad. Sci. USA 81: 8014-8018.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT editor-in-chief@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE









